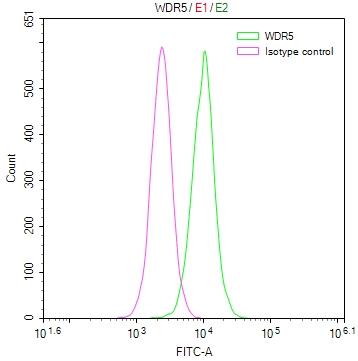
BCR Antibody
-
中文名稱:BCR兔多克隆抗體
-
貨號:CSB-PA196525
-
規格:¥1100
-
圖片:
-
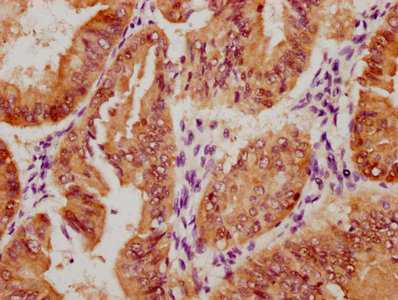
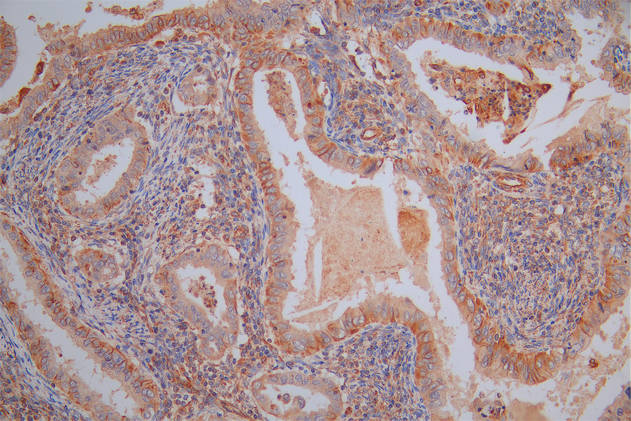
The image on the left is immunohistochemistry of paraffin-embedded Human gastic cancer tissue using CSB-PA196525(BCR Antibody) at dilution 1/15, on the right is treated with synthetic peptide. (Original magnification: ×200)
-
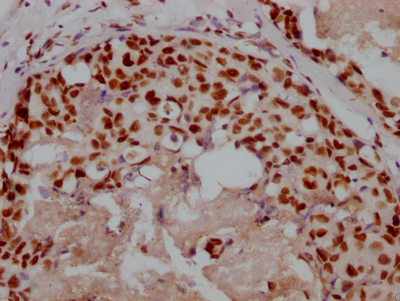
The image on the left is immunohistochemistry of paraffin-embedded Human colon cancer tissue using CSB-PA196525(BCR Antibody) at dilution 1/15, on the right is treated with synthetic peptide. (Original magnification: ×200)
-
-
其他:
產品詳情
-
Uniprot No.:
-
基因名:
-
別名:ALL antibody; bcr antibody; BCR/ABL FUSION GENE, INCLUDED antibody; BCR/FGFR1 chimera protein antibody; BCR/FGFR1 FUSION GENE, INCLUDED antibody; BCR/PDGFRA FUSION GENE, INCLUDED antibody; BCR_HUMAN antibody; BCR1 antibody; Breakpoint cluster region antibody; Breakpoint cluster region protein antibody; CML antibody; D22S11 antibody; D22S662 antibody; FGFR1/BCR chimera protein antibody; PHL antibody; Renal carcinoma antigen NY-REN-26 antibody
-
宿主:Rabbit
-
反應種屬:Human,Mouse
-
免疫原:Synthetic peptide of Human BCR
-
免疫原種屬:Homo sapiens (Human)
-
標記方式:Non-conjugated
-
抗體亞型:IgG
-
純化方式:Antigen affinity purification
-
濃度:It differs from different batches. Please contact us to confirm it.
-
保存緩沖液:-20°C, pH7.4 PBS, 0.05% NaN3, 40% Glycerol
-
產品提供形式:Liquid
-
應用范圍:ELISA,IHC
-
推薦稀釋比:
Application Recommended Dilution ELISA 1:1000-1:2000 IHC 1:25-1:100 -
Protocols:
-
儲存條件:Upon receipt, store at -20°C or -80°C. Avoid repeated freeze.
-
貨期:Basically, we can dispatch the products out in 1-3 working days after receiving your orders. Delivery time maybe differs from different purchasing way or location, please kindly consult your local distributors for specific delivery time.
-
用途:For Research Use Only. Not for use in diagnostic or therapeutic procedures.
相關產品
靶點詳情
-
功能:Protein with a unique structure having two opposing regulatory activities toward small GTP-binding proteins. The C-terminus is a GTPase-activating protein (GAP) domain which stimulates GTP hydrolysis by RAC1, RAC2 and CDC42. Accelerates the intrinsic rate of GTP hydrolysis of RAC1 or CDC42, leading to down-regulation of the active GTP-bound form. The central Dbl homology (DH) domain functions as guanine nucleotide exchange factor (GEF) that modulates the GTPases CDC42, RHOA and RAC1. Promotes the conversion of CDC42, RHOA and RAC1 from the GDP-bound to the GTP-bound form. The amino terminus contains an intrinsic kinase activity. Functions as an important negative regulator of neuronal RAC1 activity. Regulates macrophage functions such as CSF1-directed motility and phagocytosis through the modulation of RAC1 activity. Plays a major role as a RHOA GEF in keratinocytes being involved in focal adhesion formation and keratinocyte differentiation.
-
基因功能參考文獻:
- The combination of BCR-ABL1 transcript type and spleen size at diagnosis is significantly predictive for achieving an overall MMR and FFS. Incorporating these predictors could be important when making clinical decisions regarding changing therapy for CML patients treated initially with IM. PMID: 28540759
- expression of WASP inversely correlates with BCR-ABL1 levels and the progression of the disease in Chronic myeloid leukemia patients. BCR-ABL1 downregulates WASP in part by epigenetic modification of its proximal promoter. PMID: 29022901
- The imaging method achieved ultrasensitive detection of BCR/ABL fusion gene with a low detection limit down to 23 fM. And this method exhibited wide linear ranges over seven orders of magnitude and excellent discrimination ability toward target PMID: 27577607
- This is the first report evaluating the role of SOD2 in native and T351-mutated BCR-ABL-expressing cells and in a large cohort of chronic myeloid leukemia patients. In leukemic cells silenced for SOD2 expression a specific down-regulation of the expression of PRDX2 gene was found. PMID: 29550484
- The compound missense mutations in BCR-ABL kinase domain responsible to elicit disease progression, drug resistance or disease relapse in chronic myeloid leukemia. PMID: 28278078
- JNJ-26854165, an inhibitor of MDM2, inhibits proliferation and triggers cell death in a p53-independent manner in various BCR/ABL-expressing cells, which include primary leukemic cells from patients with CML blast crisis and cells expressing the Imatinib-resistant T315I BCR/ABL mutant. PMID: 27999193
- Double inhibition of the N- and C-terminal termini can disrupt Hsp90 chaperone function synergistically, but not antagonistically, in Bcr-Abl-positive human leukemia cells. PMID: 28036294
- this study identifies different BCR/Abl protein suppression patterns as a converging trait of chronic myeloid leukemia cell adaptation to energy restriction PMID: 27852045
- BCR-ABL1-positive microvesicles from chronic myeloid leukemias malignantly transform human bone marrow mesenchymal stem cells. PMID: 28836580
- Data indicate the Sp1 oncogene functions as a positive regulator for BCR/ABL expression. PMID: 27144331
- dehydrocostus lactone significantly inhibits the phosphorylation expression of Bcr/Abl, STAT5, JAK2, and STAT3 and downstream molecules including p-CrkL, Mcl-1, Bcl-XL, and Bcl-2 proteins in K562 cells. PMID: 28300289
- H19 overexpression, a frequent event in chronic myeloid leukemia, was associated with higher BCR-ABL transcript and disease progression. H19 DMR/ICR hypomethylation in CML may be one of the mechanisms mediating H19 overexpression. PMID: 28776669
- Frequent molecular monitoring and intervention are required for patients who do not show a reduction in BCR-ABL1 transcripts to these levels after stem cell transplantation. PMID: 27334764
- this study shows that BCR regulates inflammation development via the alpha subunit of casein kinase II associated with BCR PMID: 27630163
- the e13a2 BCR-ABL1 fusion transcript affects the rate, the depth, and the speed of the response to treatment with imatinib firstline, and that including the transcript type in the calculation of the baseline risk scores may improve prognostic stratification and may help the choice of the best treatment policy. PMID: 28466557
- 6 overexpression may contribute to the high proliferation and low apoptosis in chronic myeloid leukemia cells and can be regulated by BCR/ABL signal transduction through downstream phosphoinositide 3-kinase/Akt and Janus kinase/signal transducer and activator of transcription pathways, suggesting cell division cycle protein 6 as a potential therapeutic target in chronic myeloid leukemia. PMID: 28639894
- though our data support the previous findings that co-expression of BCR-ABL transcripts is due to the occurrence of exonic and intronic polymorphisms in the BCR gene, it also shows that the intronic polymorphism can arise without the linked exonic polymorphism. The occurrence of ABL kinase domain mutation is less frequent in Indian population. PMID: 27748288
- In silico three-dimensional modeling of apoptin, molecular docking experiments between apoptin model and the known structure of Bcr-Abl, and the 3D structures of SH2 domains of CrkL and Bcr-Abl, were performed. PMID: 22253690
- The present study screened for the presence of bcr-abl transcripts in the blood of a group of healthy individuals. PMID: 24535287
- Data indicate that the biosensor showed excellent analytical performance for the detection of the BCR/ABL oncogene in clinical samples of patients with leukemia. PMID: 27693719
- Studies indicate that the prognosis of BCR-ABL-positive acute myeloid leukemia (BCR-ABL+ AML) seems to depend on the cytogenetic and/or molecular background rather than on BCR-ABL itself. PMID: 27297971
- demonstrated that depletion of endogenous MAPK15 expression inhibited BCR-ABL1-dependent cell proliferation PMID: 26291129
- Identify a novel BCR-ABL/IkappaBalpha/p53 network, whereby BCR-ABL functionally inactivates a key tumor suppressor in chronic myeloid leukemia. PMID: 26295305
- Blockade of the interaction between Bcr-Abl and PTB1B by small molecule SBF-1 to overcomes imatinib-resistance of K562 cells. PMID: 26721204
- BCR-ABL1 transcript types found in Syria were similar to that of Indian Far-Eastern, African or European populations. The M-BCR rearrangement types were not dependent on white blood count, platelet count, hemoglobin level or gender of the patients. PMID: 27273956
- Data suggest elevated interleukin-1beta secretion from tyrosine kinase inhibitor- (TKI-)resistant chronic myeloid leukemia (CML) cells contributes to TKI/imatinib resistance via promotion of cell viability/migration; cells used lack BCR-ABL mutation. PMID: 26831735
- Higher incidence of BCR-ABL and lower incidence of TEL-AML1 are associated with acute lymphoblastic leukemia. PMID: 26264145
- Suggest that AIC-47 in combination with imatinib strengthened the attack on cancer energy metabolism in BCR-ABL-harboring leukemic cells. PMID: 26607903
- Molecular rearrangements and the minimal residual disease follow-up for 5 chronic myeloid leukaemia patients; 3 resulted from new rearrangements between the BCR and ABL1 sequences (the breakpoints being located within BCR exon 13 in 2 cases and within BCR exon 18 in one case). The other 2 cases revealed a complex e8-[ins]-a2 fusion transcript involving a 3rd partner gene. PMID: 26252834
- analysis of WT1 and M-BCR-ABL expressions in chronic myeloid leukemia reveals that high WT1 expression in CML patients is detected especially in the advanced stages of the disease PMID: 26429162
- BCR-ABL kinase domain mutation is associated with Philadelphia-positive leukemia. PMID: 25379619
- Separase protein levels decrease and Separase proteolytic activity increases exclusively in b3a2 p210BCR-ABL-positive cell lines under Imatinib treatment. PMID: 26087013
- our results confirm that not obtaining a BCR-ABL/ABL ratio of PMID: 25756742
- we describe a regulatory pathway modulating BCR and BCR/ABL1 expression, showing that the BCR promoter is under the transcriptional control of the MYC/MAX heterodimer. PMID: 26179066
- BCR- ABL1 mutations are associated with the clinical resistance, but may not be considered the only cause of resistance to imatinib. PMID: 25740611
- Expression profiling of adult acute lymphoblastic leukemia identifies a BCR-ABL1-like subgroup characterized by high non-response and relapse rates. PMID: 25769542
- expression of the BCR-ABL gene were confirmed by FISH, which revealed a high concordance (100%) rate. We found that real-time RT-qPCR is more reliable and should be used in Moroccan biomedical analysis laboratories to monitor CML progression PMID: 25730044
- BCR-ABL1 mutation is associated with chronic myeloid leukemia. PMID: 25721898
- Data sugget that the acquisition of additional BCR-ABL1 fusion genes in chronic myeloid leukemia (CML) in the blast phase (BP) through mitotic recombination between the derivative chromosome and the normal homologue. PMID: 26186983
- Lack of response to tyrosine kinase inhibitors associated with mutation in the BCR-ABL gene was significantly higher in imatinib-treated patients, and all mutations arose after treatment. T315I was a common treatment-emergent mutation PMID: 25615000
- High proportion of M-bcr gene is associated with chronic myeloid leukemia. PMID: 25520136
- ATRA treatment decreased DNA damage repair and suppressed acquisition of BCR-ABL mutations PMID: 24967705
- Results suggest that the EGFR and Akt pathways are involved in regulation of BCRP expression in non small cell lung carcinoma cells. PMID: 25479544
- AIC-47, acting through the PPARgamma/beta-catenin pathway, induced down-regulation of c-Myc, leading to the disruption of the bcr-abl/mTOR/hnRNP signaling pathway, and switching of the expression of PKM2 to PKM1 in acute myeloid leukemia. PMID: 25644089
- Bcr knockdown in the context of KSHV-associated disease might enhance Rac1-mediated angiogenesis. PMID: 25631082
- The epigenetic silencing of miR-23a led to derepression of BCR/ABL expression, and consequently contributes to CML development and progression. PMID: 25213664
- STAT3 is a critical signaling node in BCR-ABL1 tyrosine kinase-independent leukemia resistance that is reversed by a discovered BP-5-087. PMID: 25134459
- C817 is a promising compound for treatment of CML patients with Bcr-Abl kinase domain mutations that confer imatinib resistance. PMID: 24487968
- BCR-ABL-T315I mutation is associated with chronic myeloid leukemia. PMID: 25217883
- Leptin levels were increased in BCR-ABL p210 positive chronic myeloid leukemia patients. PMID: 25648025
顯示更多
收起更多
-
相關疾病:Leukemia, chronic myeloid (CML)
-
亞細胞定位:Cell junction, synapse, postsynaptic density. Cell projection, dendritic spine. Cell projection, axon. Cell junction, synapse.
-
數據庫鏈接:
Most popular with customers
-
-
YWHAB Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC, IF, FC
Species Reactivity: Human, Mouse, Rat
-
Phospho-YAP1 (S127) Recombinant Monoclonal Antibody
Applications: ELISA, WB, IHC
Species Reactivity: Human
-
-
-
-
-